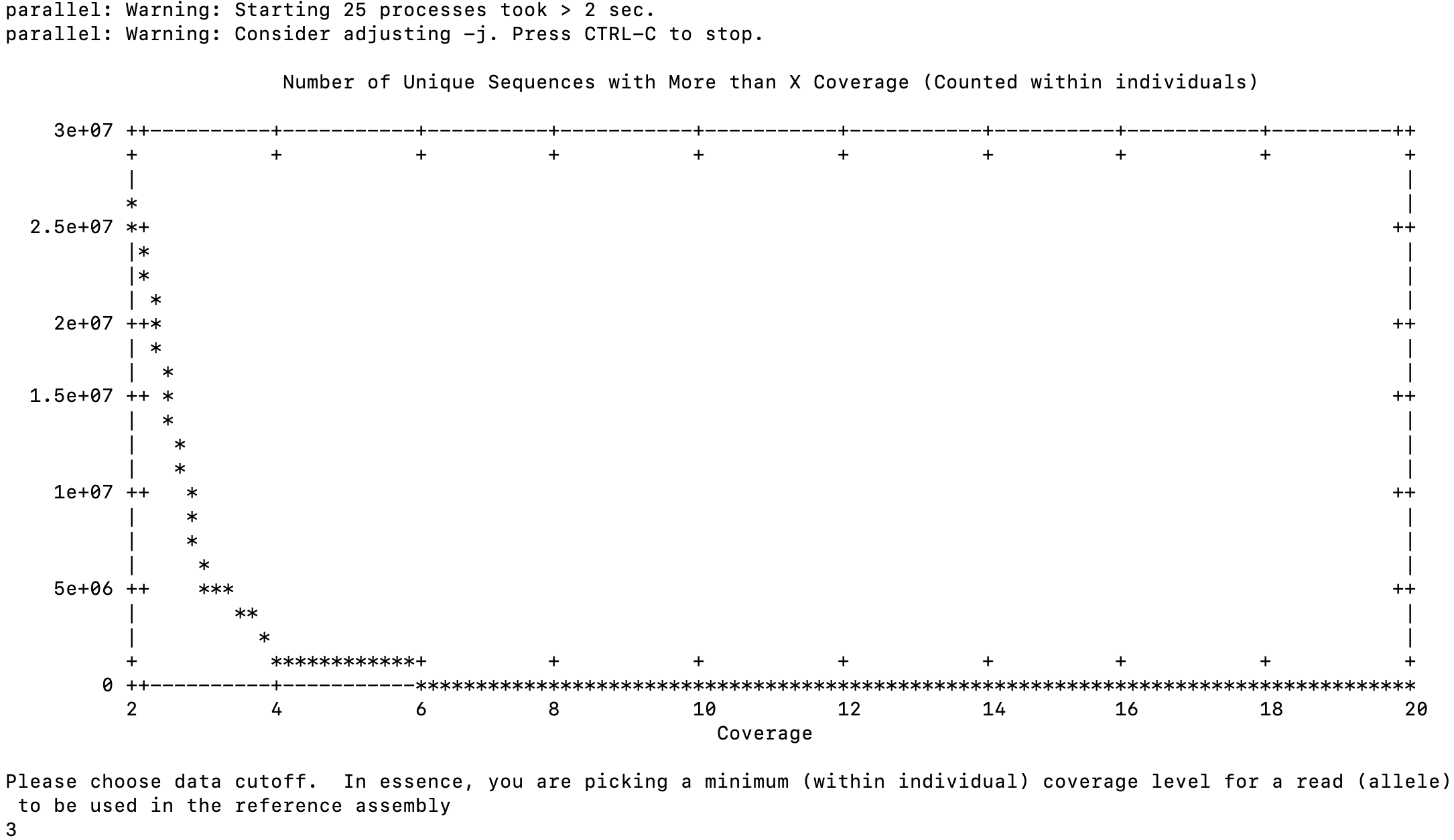
So given the output from [[dDocent All Individuals 2.5%]] and [[dDocent All Individuals 10%]], I thought I’d run a 5% representation.
screen -r 3222and sample them as:
rm(list=ls())
files <- list.files("all",pattern="F.fq.gz")
for( i in 1:length(files) ) {
file <- files[i]
tmp <- strsplit(file, split="\\.")[[1]]
ofile <- paste( paste(tmp[1],tmp[2],sep=""), tmp[3],tmp[4], sep=".")
cmd <- paste( "seqtk sample ./all/",file," 0.025 > ./all0025/", ofile, sep="")
system(cmd)
cmd <- paste("gzip ", "./all0025/",ofile,sep="")
system(cmd)
cat( format(i/length(files), digits=3), " ", ofile, "\n")
}- Started at 1040.
- 60 minute timer
- start 0.012
- end 0.264
- Estimated finish: 1500
- 60 minute timer
Trimming reduced 5% representation: - Start: Jan 31 18:51 - End: Feb 1 00:21
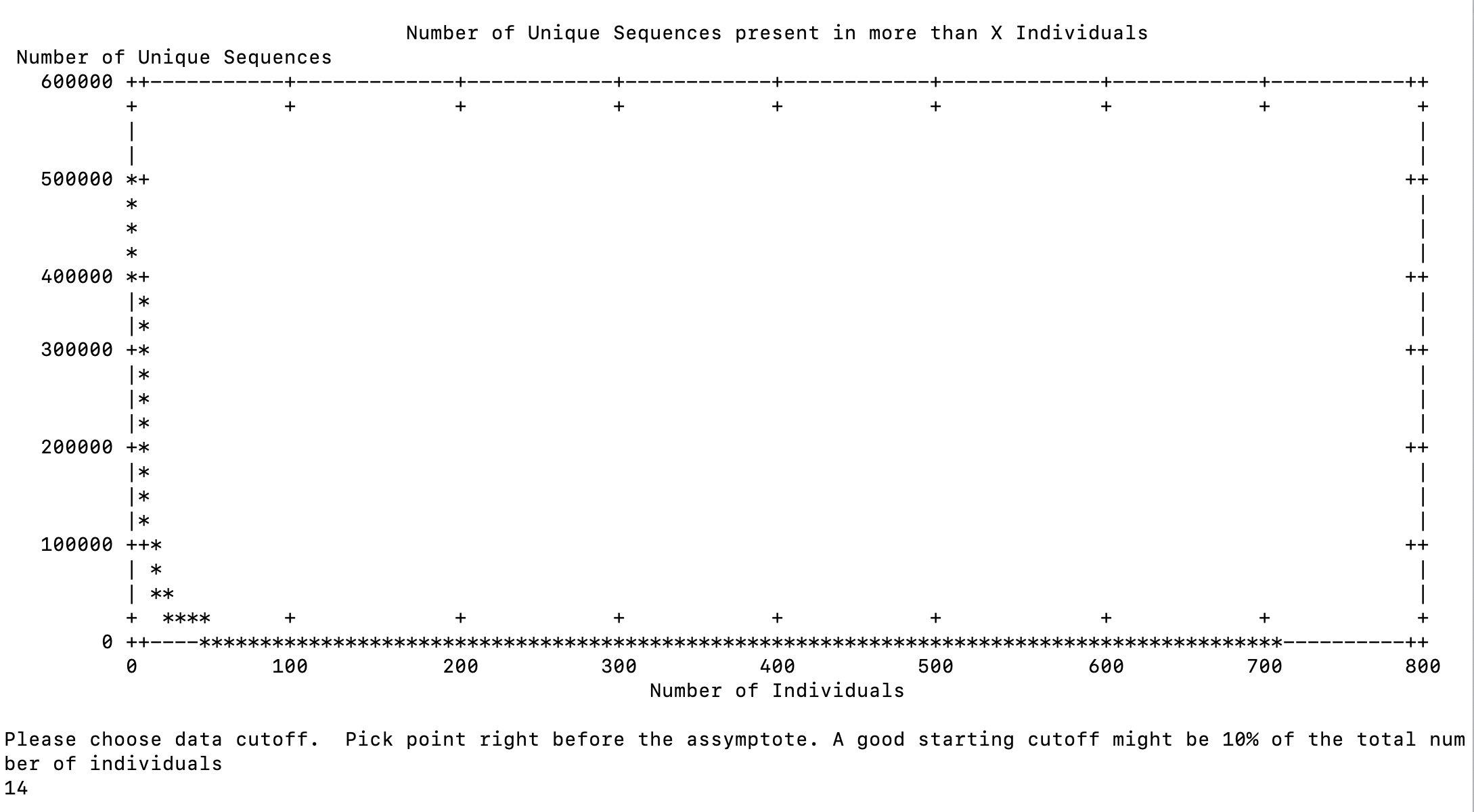
I set it to only record sequences if they are in 10% of the individuals in the full dataset.
At this point, all configuration information has been entered and dDocent may take several hours to run.
It is recommended that you move this script to a background operation and disable terminal input and output.
All data and logfiles will still be recorded.
To do this:
Press **control** and **Z** simultaneously
Type **bg** and press enter
Type **disown -h** and press enter
Now sit back, relax, and wait for your analysis to finish
dDocent assembled 88487 sequences (after cutoffs) into 57829 contigs
dDocent has finished with an analysis in /lustre/home/rjdyer/clemgu/samples/all005
dDocent started Tue Jan 31 18:21:35 EST 2023
dDocent finished Wed Feb 1 07:20:00 EST 2023