So, started again with a smaller amount of starting data. This time, I set the random sample to 2.5% of the sequences from each individual. Instructions for how I did that are here.
It took about 3 hours to do this plus another 2 hours to gzip all the data files.
dDocent Run
[rjdyer@huff10 all0025]$ dDocent
dDocent 2.9.4
Contact jpuritz@uri.edu with any problems
Checking for required software
All required software is installed!
dDocent version 2.9.4 started Mon Jan 30 13:32:43 EST 2023
1416 individuals are detected. Is this correct? Enter yes or no and press [ENTER]
yes
Proceeding with 1416 individuals
dDocent detects 256 processors available on this system.
Please enter the maximum number of processors to use for this analysis.
256
Do you want to quality trim your reads?
Type yes or no and press [ENTER]?
yes
Do you want to perform an assembly?
Type yes or no and press [ENTER].
yes
What type of assembly would you like to perform? Enter SE for single end, PE for paired-end, RPE for paired-end sequencing for RAD protocols with random shearing, or OL for paired-end sequencing that has substantial overlap.
Then press [ENTER]
SE
Reads will be assembled with Rainbow
CD-HIT will cluster reference sequences by similarity. The -c parameter (% similarity to cluster) may need to be changed for your taxa.
Would you like to enter a new c parameter now? Type yes or no and press [ENTER]
yes
Please enter new value for c. Enter in decimal form (For 90%, enter 0.9)
0.85
Do you want to map reads? Type yes or no and press [ENTER]
no
Mapping will not be performed
Do you want to use FreeBayes to call SNPs? Please type yes or no and press [ENTER]
no
Please enter your email address. dDocent will email you when it is finished running.
Dont worry; dDocent has no financial need to sell your email address to spammers.
rjdyer@vcu.edu
dDocent will require input during the assembly stage. Please wait until prompt says it is safe to move program to the background.
Trimming reads and simultaneously assembling reference sequencesRan into error
parallel: Warning: Starting 32 processes took > 2 sec.
parallel: Warning: Consider adjusting -j. Press CTRL-C to stop.Results were posted at 1725
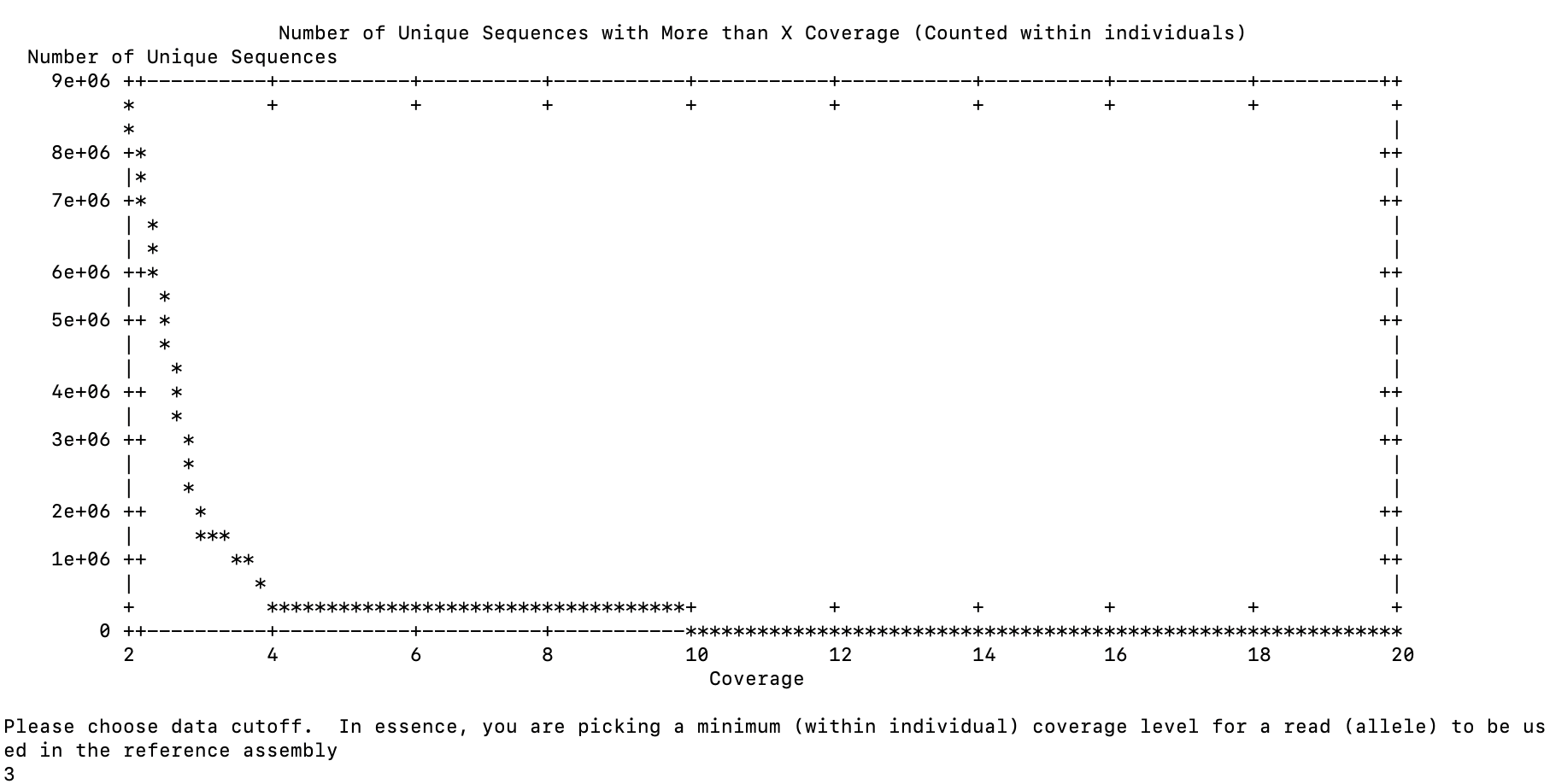
for the first question and 1528 for the next one
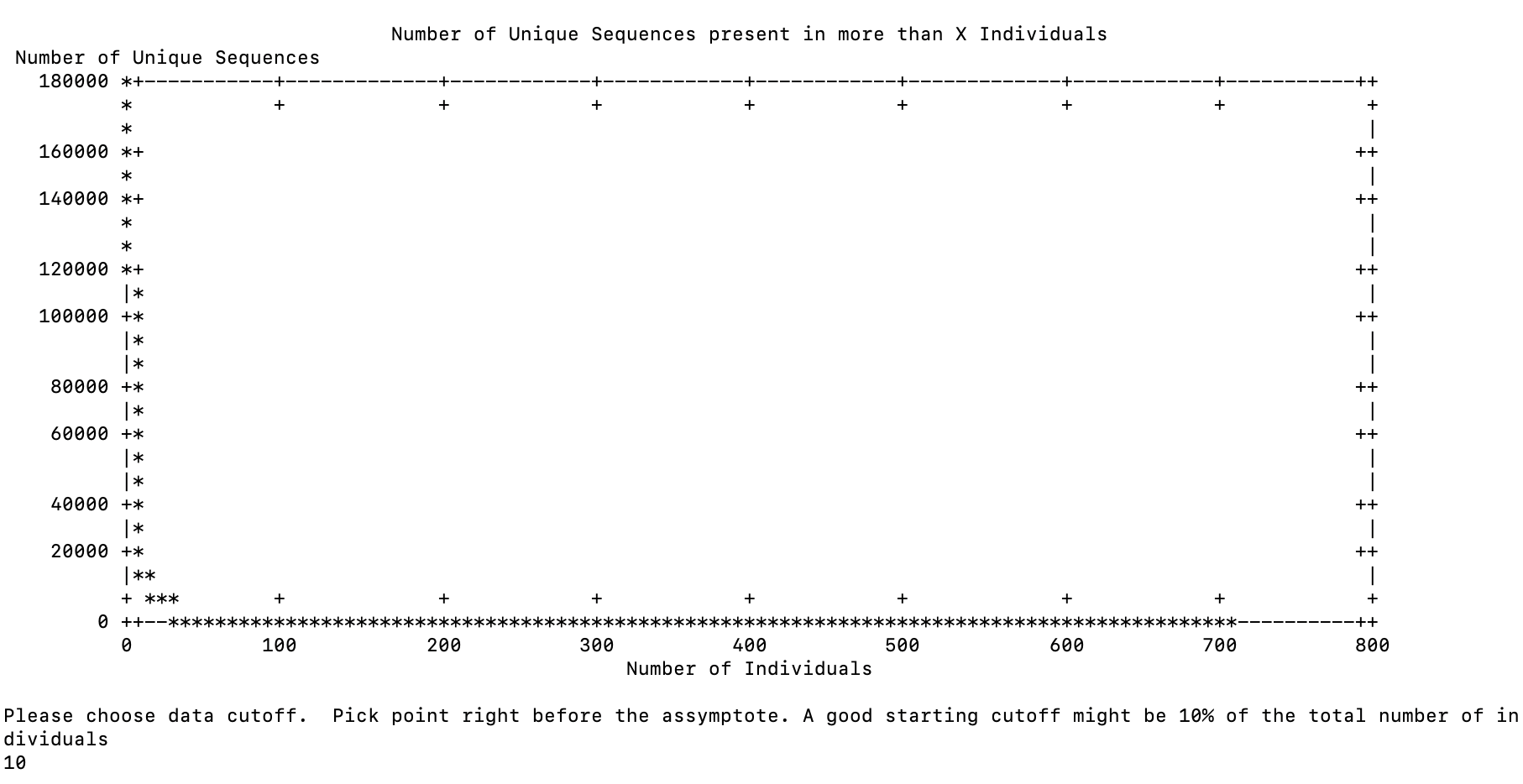
I then put it into the background and let it run.
Now sit back, relax, and wait for your analysis to finish
dDocent assembled 13966 sequences (after cutoffs) into 6676 contigs
dDocent has finished with an analysis in /lustre/home/rjdyer/clemgu/samples/all0025
dDocent started Mon Jan 30 13:32:43 EST 2023
dDocent finished Mon Jan 30 17:30:02 EST 2023